Survey of differentially methylated promoters in
prostate cancer cell lines.
Yipeng Wang1, Qiuju Yu1, Ann H Cho1, Gaelle Rondeau1, John Welsh1, Eileen Adamson2, Dan Mercola1, and Michael McClelland1
1. Sidney Kimmel Cancer Center, 10835 Road to the Cure, San Diego, California, 92121, USA
2. The Burnham Institute, Cancer Research Center, La Jolla, California, USA
Correspondence should be addressed to M McClelland, Sidney Kimmel Cancer Center, 10835 Road to the Cure, San Diego, California, 92121, USA. E-mail: mmcclelland@sdibr.org.
ABSTRACT
DNA methylation and copy number in the genomes of three immortalized prostate epithelial, and five cancer cell lines, LNCaP, PC3, PC3M, PC3M-Pro4 and PC3M-LN4, were compared using a microarray-based technique. Genomic DNA is cut with a methylation-sensitive enzyme HpaII, followed by linker ligation, PCR amplification, labeling, and hybridizion to an array of promoter sequences. Only those parts of the genomic DNA that have unmethylated restriction sites within a few hundred base pairs generate PCR products detectable on an array. Of 2732 promoter sequences on a test array, 504 (18.5%) showed differential hybridization between immortalized prostate epithelial and cancer cell lines. Among candidate hypermethylated genes in cancer-derived lines, there were eight; CD44, CDKN1A, ESR1, PLAU, RARB, SFN, TNFRSF6, and TSPY, previously observed in prostate cancer, and 13 previously known methylation targets in other cancers; ARHI, bcl-2, BRCA1, CDKN2C, GADD45A, MTAP, PGR, SLC26A4, SPARC, SYK, TJP2, UCHL1, and WIT-1. The majority of genes that appear to be both differentially methylated and differentially regulated between prostate epithelial and cancer cell lines are novel methylation targets, including PAK6, RAD50, TLX3, PIR51, MAP2K5, INSR, FBN1, GG2-1, representing a rich new source of candidate genes to study the role of DNA methylation in prostate tumors.
Table 1. Primers for methylation-specific
semi-quantitiative PCR
|
RefSeq
ID |
Gene
Symbol |
Expresssion
Ratio (log2) |
Primer Information a |
|
Putative
hypermethylated promoters in PC3M relative to 267B1 |
|||
|
NM_000082 |
CKN1 |
-0.75 |
M-FW:
GTTAATTTTCGAGAAAGGAATTAGC RW:
AAAATATCTTCAACGCCTCGAC U-FW:
ATGTTAATTTTTGAGAAAGGAATTAGTG RW:
AAAAAAAATATCTTCAACACCTCAAC |
|
NM_001008 |
RPS4Y b |
-6.36 |
M-FW:
GTTATTTAGGTTGGAGTGTAGTGGC RW:
GAATCACGAAATCAAAAAATCG U-FW:
GTTATTTAGGTTGGAGTGTAGTGGTG RW:
CAAATCACAAAATCAAAAAATCAAA |
|
NM_003118 |
SPARC b,c |
-7.16 |
M-FW:
GATATTTTCGTTTACGTCGTTAGTTC RW:
AAAAAATAAAAAAATACTCCCCCG U-FW:
GATATTTTTGTTTATGTTGTTAGTTTGT RW:
AAAAATAAAAAAATACTCCCCCAAA |
|
NM_003206 |
TCF21 |
NA |
M-FW:
AATATGTTTATCGGTTTTTTTAGCG RW:
TTAAAACTCTCCTCGATACTCTCGT U-FW:
TTTAAATATGTTTATTGGTTTTTTTAGTGA RW:
CAATTAAAACTCTCCTCAATACTCTCATT |
|
NM_003999 |
OSMR |
-2.52 |
M-FW:
ATTTTGGTTAATACGGTGAAATTTC RW:
CCAAACTAAAATACAATAACGCGAT U-FW:
TTTTGGTTAATATGGTGAAATTTTGT RW:
TCACCCAAACTAAAATACAATAACACA |
|
NM_004701 |
CCNB2 |
-1.78 |
M-FW:
GTTAAAATTTAGAGGCGTTTTACGT RW:
ACGTTTAATTATCACAACAACCGAT U-FW:
TTTTGTTAAAATTTAGAGGTGTTTTATGT RW:
CACATTTAATTATCACAACAACCAAT |
|
NM_005509 |
DMXL1 |
-1.37 |
M-FW:
ATTTCGTTTAGGGATTTGGAAATAC RW:
AAACTACAAATCCCAATATACACCG U-FW:
TTTTGTTTAGGGATTTGGAAATATG RW:
AAACTACAAATCCCAATATACACCACT |
|
NM_005732 |
RAD50 |
-2.69 |
M-FW:
ATTTTTTTGATTTTGAGATTCGC RW:
GATCCGAAACATATTTACAAACGTT U-FW:
ATTTTTTTGATTTTGAGATTTGTGG RW:
TCAATCCAAAACATATTTACAAACATT |
|
NM_005983 |
SKP2 |
-1.72 |
M-FW:
TATTTCGTGGGTCGATTAGTTTC RW:
ACTAAAAATTATAATTTCCGTCCCG U-FW:
TATTTTGTGGGTTGATTAGTTTTGT RW:
ACTAAAAATTATAATTTCCATCCCACT |
|
NM_006479 |
PIR51 |
-1.97 |
M-FW:
GTATAAATTCGGTTTTGGTGGATC RW:
CAAATTCTTATTAACTTCAACGACGA U-FW:
GTATAAATTTGGTTTTGGTGGATTG RW:
TTCTCAAATTCTTATTAACTTCAACAACA |
|
NM_014350 |
GG2-1 |
-1.94 |
M-FW:
GTTTGGAGTATTAGTGTTCGTTCG RW:
CGAAACCTTTTAAAAAAAATAAAACG U-FW:
GTTTGGAGTATTAGTGTTTGTTTGG RW:
CAAAACCTTTTAAAAAAAATAAAACAAC |
|
NM_021025 |
TLX3 |
NA |
M-FW:
GTTGTGGTTCGGGTTTTAATATTC RW:
CTACCGCAACCATTAACTACGAT U-FW:
GTTGTGGTTTGGGTTTTAATATTTG RW:
TCCTACCACAACCATTAACTACAAT |
|
NM_024501 |
HOXD1 |
-1.61 |
M-FW:
TTTTAGTGAAAGTAAGCGTCGTATC RW:
CTATCCCTCGCAATTTATAACGA U-FW:
TTTTTAGTGAAAGTAAGTGTTGTATTGG RW:
TCTTCTATCCCTCACAATTTATAACAAC |
|
NM_006142 |
SFN
c |
5.60 |
M-FW: TAAGTTGGTAGAGTAGGTCGAACGT RW: CTAAAAACAAATTTCGCTCTTCG U-FW: GGTTAAGTTGGTAGAGTAGGTTGAATG RW: CTACTAAAAACAAATTTCACTCTTCACA |
a M: primer designed to amplify methylated DNA; U: primer designed to amplify unmethylated DNA.
b Gene that does not have CpG island within the amplified promoter region.
c Gene already been known as methylation target in cancer.
Table 2. Differential amplified HpaII fragment hybridization in prostate cancer cell lines among
genes known to be methylation
targets in cancer.
|
RefSeq ID |
Gene Symbol |
Tumor Type |
Reference |
|
NM_004675 |
ARHI |
Breast cancer |
(1) |
|
NM_000633 |
bcl-2 |
Colorectal carcinoma |
(2) |
|
NM_007296 |
BRCA1 |
Breast cancer Ovarian cancer Cervical cancer |
(3-5) |
|
NM_000610 |
CD44 |
Prostate cancer Colorectal
cancer Neuroblastoma Gastric
cancer |
(6-13) |
|
NM_000389 |
CDKN1A |
Prostate cancer Lymphoma Leukemia |
(14-17) |
|
NM_001262 |
CDKN2C |
Hodgkin lymphomas |
(18). |
|
NM_000125 |
ESR1 |
Prostate cancer Colorectal cancer Breast cancer Lung cancer |
(3,19-22) |
|
NM_001924 |
GADD45A |
Breast cancer |
(23) |
|
NM_002451 |
MTAP |
Malignant melanoma |
(24) |
|
NM_000926 |
PGR |
Breast cancer, Cervical cancer |
(25,26) |
|
NM_002658 |
PLAU |
Prostate cancer Breast cancer |
(27,28) |
|
NM_000965 |
RARB |
Prostate cancer Testicular lymphoma Cervical cancer Breast Cancer colorectal cancers |
(29-33) |
|
NM_006142 |
SFN |
Prostate cancer Overian cancer Skin cancer Lung cancer Oral cancer Vulval cancer Gastric cancer Breast cancer |
(34-41) |
|
NM_000441 |
SLC26A4 |
Thyroid tumorigenesis |
(42) |
|
NM_003118 |
SPARC |
Pancreatic cancer |
(43) |
|
NM_003177 |
SYK |
Breast cancer Gastric cancer Overian cancer T-lineage acute lymphoblastic leukemia |
(44-47) |
|
NM_004817 |
TJP2 |
Pancreatic cancer |
(48) |
|
NM_000043 |
TNFRSF6 |
Prostate cancer Bladder cancer |
(49) |
|
NM_003308 |
TSPY |
Prostate cancer |
(50) |
|
NM_004181 |
UCHL1 |
Pancreatic cancer |
(48) |
|
NM_015855 |
WIT-1 |
Acute myeloid leukemia |
(51) |
References in
Table 2
1. Yuan, J, Luo, RZ, Fujii, S, Wang, L, Hu, W, Andreeff, M, Pan, Y, Kadota, M, Oshimura, M, Sahin, AA et al. (2003). Aberrant methylation and silencing of ARHI, an imprinted tumor suppressor gene in which the function is lost in breast cancers Cancer Res, 63, 4174-4180.
2. Babidge, WJ, Butler, LM, Burton, MA and Cowled, PA. (2001). Methylation of CpG sites in exon 2 of the bcl-2 gene occurs in colorectal carcinoma Anticancer Res, 21, 2809-2814.
3. Parrella, P, Poeta, ML, Gallo, AP, Prencipe, M, Scintu, M, Apicella, A, Rossiello, R, Liguoro, G, Seripa, D, Gravina, C et al. (2004). Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors Clin Cancer Res, 10, 5349-5354.
4. Narayan, G, Arias-Pulido, H, Nandula, SV, Basso, K, Sugirtharaj, DD, Vargas, H, Mansukhani, M, Villella, J, Meyer, L, Schneider, A et al. (2004). Promoter hypermethylation of FANCF: disruption of Fanconi Anemia-BRCA pathway in cervical cancer Cancer Res, 64, 2994-2997.
5. Wang, C, Horiuchi, A, Imai, T, Ohira, S, Itoh, K, Nikaido, T, Katsuyama, Y and Konishi, I. (2004). Expression of BRCA1 protein in benign, borderline, and malignant epithelial ovarian neoplasms and its relationship to methylation and allelic loss of the BRCA1 gene J Pathol, 202, 215-223.
6. Kito, H, Suzuki, H, Ichikawa, T, Sekita, N, Kamiya, N, Akakura, K, Igarashi, T, Nakayama, T, Watanabe, M, Harigaya, K et al. (2001). Hypermethylation of the CD44 gene is associated with progression and metastasis of human prostate cancer Prostate, 49, 110-115.
7. Lou, W, Krill, D, Dhir, R, Becich, MJ, Dong, JT, Frierson, HF, Jr., Isaacs, WB, Isaacs, JT and Gao, AC. (1999). Methylation of the CD44 metastasis suppressor gene in human prostate cancer Cancer Res, 59, 2329-2331.
8. Verkaik, NS, Trapman, J, Romijn, JC, Van der Kwast, TH and Van Steenbrugge, GJ. (1999). Down-regulation of CD44 expression in human prostatic carcinoma cell lines is correlated with DNA hypermethylation Int J Cancer, 80, 439-443.
9. Verkaik, NS, van Steenbrugge, GJ, van Weerden, WM, Bussemakers, MJ and van der Kwast, TH. (2000). Silencing of CD44 expression in prostate cancer by hypermethylation of the CD44 promoter region Lab Invest, 80, 1291-1298.
10. Woodson, K, Hayes, R, Wideroff, L, Villaruz, L and Tangrea, J. (2003). Hypermethylation of GSTP1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites Prostate, 55, 199-205.
11. Stallmach, A, Wittig, BM, Kremp, K, Goebel, R, Santourlidis, S, Zeitz, M, Menges, M, Raedle, J, Zeuzem, S and Schulz, WA. (2003). Downregulation of CD44v6 in colorectal carcinomas is associated with hypermethylation of the CD44 promoter region Exp Mol Pathol, 74, 262-266.
12. Yan, P, Muhlethaler, A, Bourloud, KB, Beck, MN and Gross, N. (2003). Hypermethylation-mediated regulation of CD44 gene expression in human neuroblastoma Genes Chromosomes Cancer, 36, 129-138.
13. Sato, S, Yokozaki, H, Yasui, W, Nikai, H and Tahara, E. (1999). Silencing of the CD44 gene by CpG methylation in a human gastric carcinoma cell line Jpn J Cancer Res, 90, 485-489.
14. Konishi, N, Nakamura, M, Kishi, M, Nishimine, M, Ishida, E and Shimada, K. (2002). DNA hypermethylation status of multiple genes in prostate adenocarcinomas Jpn J Cancer Res, 93, 767-773.
15. Konishi, N, Nakamura, M, Kishi, M, Nishimine, M, Ishida, E and Shimada, K. (2002). Heterogeneous methylation and deletion patterns of the INK4a/ARF locus within prostate carcinomas Am J Pathol, 160, 1207-1214.
16. Go, JH. (2003). Methylation analysis of cyclin-dependent kinase inhibitor genes in primary gastrointestinal lymphomas Mod Pathol, 16, 752-755.
17. Roman-Gomez, J, Castillejo, JA, Jimenez, A, Gonzalez, MG, Moreno, F, Rodriguez Mdel, C, Barrios, M, Maldonado, J and Torres, A. (2002). 5' CpG island hypermethylation is associated with transcriptional silencing of the p21(CIP1/WAF1/SDI1) gene and confers poor prognosis in acute lymphoblastic leukemia Blood, 99, 2291-2296.
18. Sanchez-Aguilera, A, Delgado, J, Camacho, FI, Sanchez-Beato, M, Sanchez, L, Montalban, C, Fresno, MF, Martin, C, Piris, MA and Garcia, JF. (2004). Silencing of the p18INK4c gene by promoter hypermethylation in Reed-Sternberg cells in Hodgkin lymphomas Blood, 103, 2351-2357.
19. Yegnasubramanian, S, Kowalski, J, Gonzalgo, ML, Zahurak, M, Piantadosi, S, Walsh, PC, Bova, GS, De Marzo, AM, Isaacs, WB and Nelson, WG. (2004). Hypermethylation of CpG islands in primary and metastatic human prostate cancer Cancer Res, 64, 1975-1986.
20. Li, LC, Shiina, H, Deguchi, M, Zhao, H, Okino, ST, Kane, CJ, Carroll, PR, Igawa, M and Dahiya, R. (2004). Age-dependent methylation of ESR1 gene in prostate cancer Biochem Biophys Res Commun, 321, 455-461.
21. Belshaw, NJ, Elliott, GO, Williams, EA, Bradburn, DM, Mills, SJ, Mathers, JC and Johnson, IT. (2004). Use of DNA from human stools to detect aberrant CpG island methylation of genes implicated in colorectal cancer Cancer Epidemiol Biomarkers Prev, 13, 1495-1501.
22. Marchevsky, AM, Tsou, JA and Laird-Offringa, IA. (2004). Classification of individual lung cancer cell lines based on DNA methylation markers: use of linear discriminant analysis and artificial neural networks J Mol Diagn, 6, 28-36.
23. Wang, W, Huper, G, Guo, Y, Murphy, SK, Olson, JA and Marks, JR. (2005). Analysis of methylation-sensitive transcriptome identifies GADD45a as a frequently methylated gene in breast cancer Oncogene, 24, 2705-2714.
24. Behrmann, I, Wallner, S, Komyod, W, Heinrich, PC, Schuierer, M, Buettner, R and Bosserhoff, AK. (2003). Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma Am J Pathol, 163, 683-690.
25. Widschwendter, A, Muller, HM, Fiegl, H, Ivarsson, L, Wiedemair, A, Muller-Holzner, E, Goebel, G, Marth, C and Widschwendter, M. (2004). DNA methylation in serum and tumors of cervical cancer patients Clin Cancer Res, 10, 565-571.
26. Widschwendter, M, Siegmund, KD, Muller, HM, Fiegl, H, Marth, C, Muller-Holzner, E, Jones, PA and Laird, PW. (2004). Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen Cancer Res, 64, 3807-3813.
27. Xing, RH and Rabbani, SA. (1999). Transcriptional regulation of urokinase (uPA) gene expression in breast cancer cells: role of DNA methylation Int J Cancer, 81, 443-450.
28. Pakneshan, P, Xing, RH and Rabbani, SA. (2003). Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo Faseb J, 17, 1081-1088.
29. Yamamoto, H, Min, Y, Itoh, F, Imsumran, A, Horiuchi, S, Yoshida, M, Iku, S, Fukushima, H and Imai, K. (2002). Differential involvement of the hypermethylator phenotype in hereditary and sporadic colorectal cancers with high-frequency microsatellite instability Genes Chromosomes Cancer, 33, 322-325.
30. Farias, EF, Arapshian, A, Bleiweiss, IJ, Waxman, S, Zelent, A and Mira, YLR. (2002). Retinoic acid receptor alpha2 is a growth suppressor epigenetically silenced in MCF-7 human breast cancer cells Cell Growth Differ, 13, 335-341.
31. Narayan, G, Arias-Pulido, H, Koul, S, Vargas, H, Zhang, FF, Villella, J, Schneider, A, Terry, MB, Mansukhani, M and Murty, VV. (2003). Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome Mol Cancer, 2, 24.
32. Kawakami, T, Okamoto, K, Kataoka, A, Koizumi, S, Iwaki, H, Sugihara, H, Reeve, AE, Ogawa, O and Okada, Y. (2003). Multipoint methylation analysis indicates a distinctive epigenetic phenotype among testicular germ cell tumors and testicular malignant lymphomas Genes Chromosomes Cancer, 38, 97-101.
33. Singal, R, Ferdinand, L, Reis, IM and Schlesselman, JJ. (2004). Methylation of multiple genes in prostate cancer and the relationship with clinicopathological features of disease Oncol Rep, 12, 631-637.
34. Urano, T, Takahashi, S, Suzuki, T, Fujimura, T, Fujita, M, Kumagai, J, Horie-Inoue, K, Sasano, H, Kitamura, T, Ouchi, Y et al. (2004). 14-3-3sigma is down-regulated in human prostate cancer Biochem Biophys Res Commun, 319, 795-800.
35. Kaneuchi, M, Sasaki, M, Tanaka, Y, Shiina, H, Verma, M, Ebina, Y, Nomura, E, Yamamoto, R, Sakuragi, N and Dahiya, R. (2004). Expression and methylation status of 14-3-3 sigma gene can characterize the different histological features of ovarian cancer Biochem Biophys Res Commun, 316, 1156-1162.
36. Lodygin, D, Yazdi, AS, Sander, CA, Herzinger, T and Hermeking, H. (2003). Analysis of 14-3-3sigma expression in hyperproliferative skin diseases reveals selective loss associated with CpG-methylation in basal cell carcinoma Oncogene, 22, 5519-5524.
37. Osada, H, Tatematsu, Y, Yatabe, Y, Nakagawa, T, Konishi, H, Harano, T, Tezel, E, Takada, M and Takahashi, T. (2002). Frequent and histological type-specific inactivation of 14-3-3sigma in human lung cancers Oncogene, 21, 2418-2424.
38. Gasco, M, Bell, AK, Heath, V, Sullivan, A, Smith, P, Hiller, L, Yulug, I, Numico, G, Merlano, M, Farrell, PJ et al. (2002). Epigenetic inactivation of 14-3-3 sigma in oral carcinoma: association with p16(INK4a) silencing and human papillomavirus negativity Cancer Res, 62, 2072-2076.
39. Gasco, M, Sullivan, A, Repellin, C, Brooks, L, Farrell, PJ, Tidy, JA, Dunne, B, Gusterson, B, Evans, DJ and Crook, T. (2002). Coincident inactivation of 14-3-3sigma and p16INK4a is an early event in vulval squamous neoplasia Oncogene, 21, 1876-1881.
40. Umbricht, CB, Evron, E, Gabrielson, E, Ferguson, A, Marks, J and Sukumar, S. (2001). Hypermethylation of 14-3-3 sigma (stratifin) is an early event in breast cancer Oncogene, 20, 3348-3353.
41. Suzuki, H, Itoh, F, Toyota, M, Kikuchi, T, Kakiuchi, H and Imai, K. (2000). Inactivation of the 14-3-3 sigma gene is associated with 5' CpG island hypermethylation in human cancers Cancer Res, 60, 4353-4357.
42. Xing, M, Tokumaru, Y, Wu, G, Westra, WB, Ladenson, PW and Sidransky, D. (2003). Hypermethylation of the Pendred syndrome gene SLC26A4 is an early event in thyroid tumorigenesis Cancer Res, 63, 2312-2315.
43. Sato, N, Fukushima, N, Maehara, N, Matsubayashi, H, Koopmann, J, Su, GH, Hruban, RH and Goggins, M. (2003). SPARC/osteonectin is a frequent target for aberrant methylation in pancreatic adenocarcinoma and a mediator of tumor-stromal interactions Oncogene, 22, 5021-5030.
44. Yuan, Y, Liu, H, Sahin, A and Dai, JL. (2004). Reactivation of SYK expression by inhibition of DNA methylation suppresses breast cancer cell invasiveness Int J Cancer.
45. Wang, S, Ding, YB, Chen, GY, Xia, JG and Wu, ZY. (2004). Hypermethylation of Syk gene in promoter region associated with oncogenesis and metastasis of gastric carcinoma World J Gastroenterol, 10, 1815-1818.
46. Dhillon, VS, Young, AR, Husain, SA and Aslam, M. (2004). Promoter hypermethylation of MGMT, CDH1, RAR-beta and SYK tumour suppressor genes in granulosa cell tumours (GCTs) of ovarian origin Br J Cancer, 90, 874-881.
47. Goodman, PA, Burkhardt, N, Juran, B, Tibbles, HE and Uckun, FM. (2003). Hypermethylation of the spleen tyrosine kinase promoter in T-lineage acute lymphoblastic leukemia Oncogene, 22, 2504-2514.
48. Sato, N, Fukushima, N, Maitra, A, Matsubayashi, H, Yeo, CJ, Cameron, JL, Hruban, RH and Goggins, M. (2003). Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays Cancer Res, 63, 3735-3742.
49. Santourlidis, S, Warskulat, U, Florl, AR, Maas, S, Pulte, T, Fischer, J, Muller, W and Schulz, WA. (2001). Hypermethylation of the tumor necrosis factor receptor superfamily 6 (APT1, Fas, CD95/Apo-1) gene promoter at rel/nuclear factor kappaB sites in prostatic carcinoma Mol Carcinog, 32, 36-43.
50. Dasari, VK, Deng, D, Perinchery, G, Yeh, CC and Dahiya, R. (2002). DNA methylation regulates the expression of Y chromosome specific genes in prostate cancer J Urol, 167, 335-338.
51. Plass, C, Yu, F, Yu, L, Strout, MP, El-Rifai, W, Elonen, E, Knuutila, S, Marcucci, G, Young, DC, Held, WA et al. (1999). Restriction landmark genome scanning for aberrant methylation in primary refractory and relapsed acute myeloid leukemia; involvement of the WIT-1 gene Oncogene, 18, 3159-3165.
Table 3. Promoters that show large differences in both HpaII fragment amplficiation and RNA
expression, between PC3M and 267B1
|
RefSeq ID |
Gene Symbol |
Gene Full Name |
Expresssion Ratio a |
|
Genes with reduced HpaII fragment hybridization in PC3M
(candidate hypermethylated genes) |
|||
|
NM_000138 NM_000546 NM_000985 NM_001008 NM_001790 NM_002082 NM_002749 NM_003118 NM_003714 NM_003999 NM_004472 NM_004663 NM_004701 NM_005509 NM_005732 NM_005983 NM_006282 NM_006479 NM_012382 NM_014621 NM_018163 NM_018268 NM_024501 NM_024558 NM_024796 NM_033028 NM_133338 NM_006194 NM_053001 |
FBN1 TP53 RPL17 RPS4Y b CDC25C GPRK6 MAPK7 SPARC b STC2 OSMR FOXD1 b RAB11A CCNB2 DMXL1 RAD50 SKP2 STK4 PIR51 OSRF HOXD4 FLJ10634 FLJ10904 HOXD1 C14orf138 FLJ22639 BBS4 RAD17 PAX9 c OSR2 c |
fibrillin 1 tumor protein p53 ribosomal protein L17 ribosomal protein S4,
Y-linked Y isoform cell division cycle 25C
protein, isoform a G protein-coupled receptor
kinase 6 mitogen-activated protein
kinase 7 isoform 1 secreted protein, acidic,
cysteine-rich (osteonectin) stanniocalcin 2 oncostatin M receptor forkhead box D1 RAB11A, member RAS oncogene
family cyclin B2 Dmx-like 1 RAD50 homolog isoform 1 S-phase kinase-associated
protein 2 isoform 1 serine/threonine kinase 4 RAD51-interacting protein osmosis responsive factor homeo box D4 hypothetical protein
FLJ10634 hypothetical protein
FLJ10904 homeo box D1 hypothetical protein
FLJ13920 hypothetical protein
FLJ22639 Bardet-Biedl syndrome 4 RAD17 homolog isoform 1 paired box gene 9 odd-skipped-related 2A
protein |
-5.21 -4.66 -1.17 -6.36 -2.29 -1.76 -1.62 -4.28 -1.42 -2.52 -2.68 -1.13 -1.78 -1.37 -2.69 -1.47 -1.36 -1.97 -2.36 -3.07 -1.66 -4.17 -1.61 -1.56 -1.00 -1.54 -1.63 2.55 1.43 |
|
Genes with increased HpaII fragment hybridization in PC3M
(candidate hypomethylated genes) |
|||
|
NM_001123 NM_002290 NM_002467 NM_002658 NM_004693 NM_005555 NM_006142 NM_030759 NM_032804 NM_018649 NM_020177 |
ADK LAMA4 MYC PLAU K6HF b KRT6B b SFN NRBF-2 FLJ14547 H2AFY2 c FEM1C c |
adenosine kinase isoform a laminin, alpha 4 precursor v-myc myelocytomatosis viral
oncogene homolog plasminogen activator,
urokinase cytokeratin type II keratin 6B stratifin nuclear receptor binding
factor-2 hypothetical protein
FLJ14547 core histone macroH2A2.2 feminization 1 homolog a |
1.69 2.11 2.51 1.75 9.27 1.10 5.60 1.45 2.39 -2.02 -1.30 |
a Ratios: log2(PC3M/267B1)
b Promoter does not have CpG island within the amplified promoter region.
c Promoter where HpaII fragment hybridization was not correlated with RNA expression level.
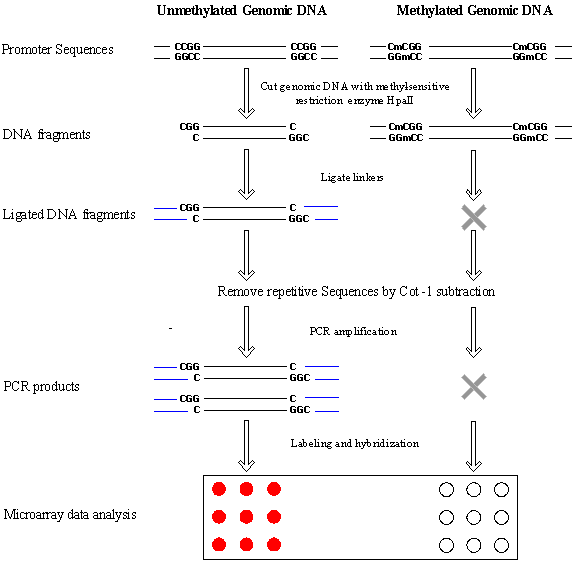
Figure 1. Schematic
of the protocol for detecting differences in HpaII fragment amplification between samples.
Figure
2. Estimation of data reproducibility and significance of differences.
M: log base 2 ratio of each spot
after print-tip loess normalization and scale between-array normalization. A:
average of two channels’ log intentisies of each spot; a measurement of the
overall brightness of the spot. All data involes at least six arrays. The p
value is for the moderated t-test. Figure 2A: M-A plot, hybridization of
amplified HpaII fragments from 267B1
vs. 267B1; Figure 2B: M-p plot, hybridization of amplified HpaII fragments from 267B1 vs. 267B1.
Figure 2C: M-A plot, hybridization of amplified HpaII fragments from PC3M vs. 267B1; Figure 2D: M-p plot, hybridization of amplified HpaII fragments from PC3M vs. 267B1.
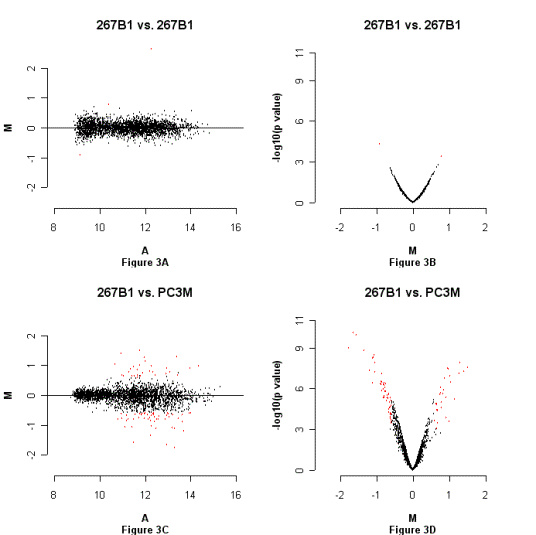
Figure 3. Detection of DNA methylation
changes using methylation-specific semi-quantitative PCR. Fourteen
promoters that displayed possible differential methylation in the array assay
between 267B1 and PC3M (see Table 1) were investigated by
methylation-specific semi-quantative PCR. The proportion of methylation for
each promoter is calculated.
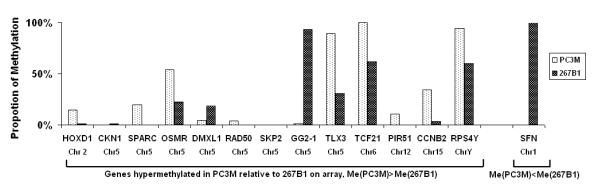
Figure 4.
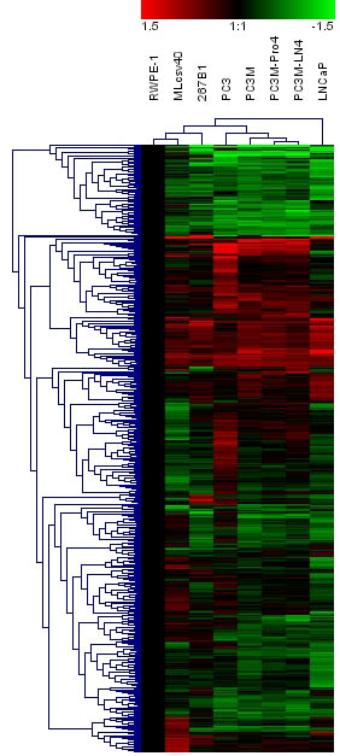
Hierarchical Cluster of hybridized amplified HpaII fragments for
eight cell lines. Figure 4A: 504 promoters that are statistical differentially
hybridized between at least one of the five prostate cancer cell lines (LNCaP,
PC3, PC3M, PC3M-Pro4, PC3M-LN4) and at least one of the three relative normal
prostate cell lines (RWPE-1, 267B1, Mlcsv40) are shown. The normalized
hybridization ratios of these cell lines relative to RWPE-1 were used for
hierarchical clustering. Red indicates higher HpaII fragment hybridization relative to
RWPE-1, which usually indicates less methylation or a higher copy number. Green indicates lower hybridization, which usually
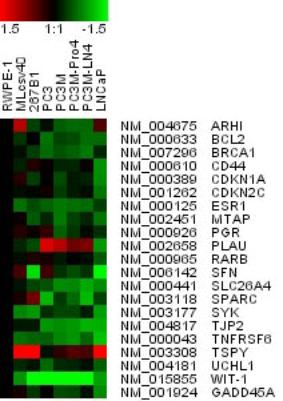
indicates greater methylation or a lower copy number. Figure 4B: Clustering
for 21 genes known to be regulated in cancer.
|
Figure
4A |
Figure
4B |
|
|
|
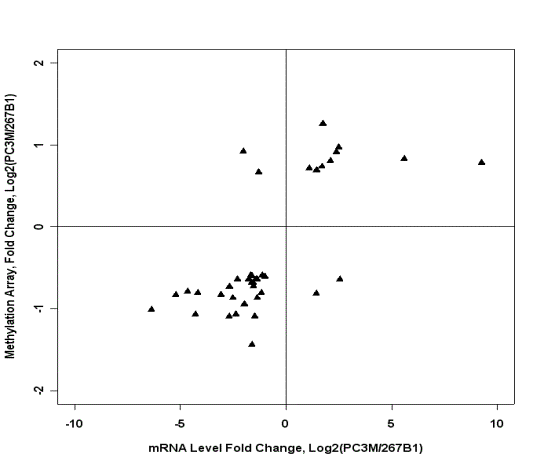
Figure 5. Comparison of amplified HpaII fragment data to Affymetrix RNA expression data. Decreases in signal from HpaII fragments (which is usually due to an increase in DNA methylation) between cell lines are generally associated with a decrease in RNA expression. The upper right quadrant contains genes with higher RNA expression in PC3M than in 267B1 and higher yield of HpaII fragments, in PC3M. The lower left quadrant contains genes with lower RNA expression in PC3M than in 267B1 and also lower yield of HpaII fragment hybridization in PC3M.
Figure
6. Effects of methylation inhibitor (5-aza-2’-deoxycytidine, DAC)
on methylation status of LNCaP.
M-p plots, HpaII fragment hybridization pattern of LNCaP before and
after treated with DAC. M: log base 2 ratio of each spot after composite normalization and scale
between-array normalization. The p value is from a moderated t-test. Figure 6A: M-p plot for all promoters; Figure 6B: M-p plot for 191 promoters
putatively hyper-methylated in LNCaP relative to at least one of the three
normal prostate cell lines.
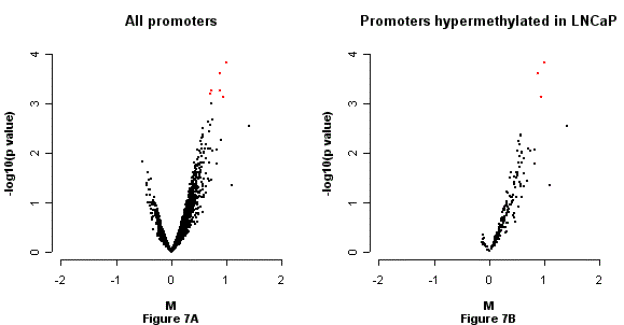
Figure 7. DNA copy number changes measured by CGH on
promoter array. Figure 7A-C: normalized
HpaII fragment hybridization ratios
for three different prostate cancer cell lines compared to 267B1 plotted
against the relative chromosomal position of each promoter. Figure 7A:
LNCaP; Figure 7B: PC3; Figure 7C: PC3M. Figure 7D: PC3M (MspI-ligation-PCR),
compared to 267B1 (MspI-ligation-PCR), against chromosomal position; Figure 7E: PC3M (RNA expression
level), compared to 267B1 RNA expression level, against chromosomal
position.

Supplement A.
Differential hybridization of amplified HpaII frangments between prostate
cancer cell lines.
Supplement B. Promoter Array Gene List.
Html Version
Excel Version
Text Version
Supplement Table W1.
Html Version
Excel Version
Text Version